

- SEARCH NUCLEOTIDE SEQUENCE ON CLC MAIN WORKBENCH SOFTWARE
- SEARCH NUCLEOTIDE SEQUENCE ON CLC MAIN WORKBENCH DOWNLOAD
- SEARCH NUCLEOTIDE SEQUENCE ON CLC MAIN WORKBENCH FREE
UGENE: high performance genome analysis suite Proceedings of the Fifth Moscow International Congress on Biotechnology 2():405–406.Advancements in high-throughput nucleotide sequencing techniques have brought with them state-of-the-art bioinformatics programs and software packages. UGENE: interactive computational schemes for genome analysis Proceedings of the Fifth Moscow International Congress on Biotechnology 3():14–15. Multitasking software system for DNA analysis Proceedings of the Sixth International Conference on Bioinformatics of Genome Regulation and Structure 1():78.

SEARCH NUCLEOTIDE SEQUENCE ON CLC MAIN WORKBENCH FREE
Free open source bioinformatics projects. UGENE was awarded "The best FOSS project in Russia – 2010" in the category "Group project" at the Linux Format magazine contest. The features to be included into the next release are mostly initiated by users. SEARCH NUCLEOTIDE SEQUENCE ON CLC MAIN WORKBENCH DOWNLOAD
One can also download a development snapshot of the software. By the end of iteration a release comes out. UGENE is primarily developed by Unipro LLC.
Other formats: Bairoch ( enzymes info), HMM ( HMMER profiles), PWM and PFM ( position matrices), etc. Short reads: Sequence Alignment/Map(SAM) (.sam), ACE (.ace), FASTQ (.fastq). Multiple sequence alignments: Clustal (.aln), MSF (.msf), Stockholm (.sto), Nexus (.nex). Sequences and annotations: FASTA (.fa), GenBank (.gb), EMBL (.emb), GFF(.gff). The results are saved as a set of annotations to a specified file in the GenBank format. UGENE Query Designer allows a user to analyze a nucleotide sequence using different algorithms ( Repeats finder, ORF finder, Weight matrix matching, etc.) at the same time imposing constraints on the positional relationship of the results obtained from the algorithms.Ī schema of the algorithms and constraints is either created from the GUI or edited as a plain text. The workflow schemas can be run both locally and remotely, either using the graphical interface or launched from the command line. Using the Workflow Designer one can also create custom workflow elements. The elements that a schema consists of correspond to the bulk of algorithms integrated into UGENE. UGENE Workflow Designer allows creating and running complex computational workflow schemas. The Alignment editor is used to visualize, analyze and modify a nucleic acid or protein multiple sequence alignment. Depending on the sequence type and the options selected the followings views can be presented inside the Sequence view window:Ģ. 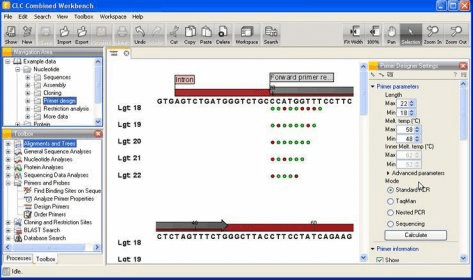
The Sequence view is used to visualize, analyze and modify nucleic acid or protein sequences. The software has three main views to display biological data on the user’s screen.ġ.
Search for a pattern of various algorithms’ results in a nucleic acid sequence with UGENE Query Designer. Combining various algorithms into custom workflows with UGENE Workflow Designer. Local sequence alignment with optimized Smith-Waterman algorithm. Building (using integrated PHYLIP package) and viewing phylogenetic trees. Protein secondary structure prediction with GOR IV and PSIPRED algorithms. 3D structure viewer for files in PDB and MMDB formats, anaglyph view support. Aligning short reads with Bowtie and UGENE genome aligner. Search for transcription factor binding sites ( TFBS) with weight matrix and SITECON algorithms. Constructing dotplots for nucleic acid sequences. Search for direct, inverted and tandem repeats in DNA sequences. Integrated Primer3 package for PCR primers design. Restriction analysis with integrated REBASE restriction enzyme database. Multiple sequence alignment: Clustal, MUSCLE, Kalign, MAFFT, T-Coffee. Search through online databases: NCBI, PDB, UniProtKB/Swiss-Prot, UniProtKB/TrEMBL. Creating, editing and annotating nucleic acid and protein sequences. The software supports the following features:







 0 kommentar(er)
0 kommentar(er)
